GEO599/CS519 Ecosystem Informatics Winter 2005 Assignment 2
In this assignment, you will learn how to apply BUGS to perform
some simple data analysis of data on human heights.
Obtaining WinBUGS
Please execute the following steps:
- Download and install WinBUGS from the BUGS website.
- Download the 2005 WinBUGS license key. Examine this
file in a text editor and follow the instructions therein for loading
the key into WinBUGS.
Running the Fully Observed Example
Now we will use WinBUGS to repeat (and improve upon) the example that
I did in class where we compared the height of men and women in the
case where we observe both the sex and the height of each data point.
- In WinBUGS, open Help > Doodle Help and consult it while you
create a model for the sex-height data analysis problem. The model is
shown in the image below:
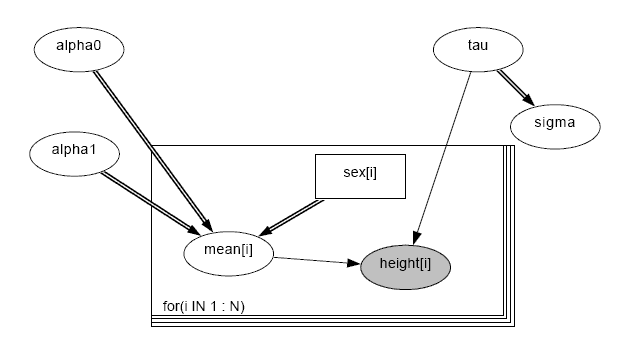
Each node can be one of three kinds: stochastic (a random variable),
logical (computed deterministically via an algebraic formula), and
constant (specified in the data file). In this model,
sex[i] should be constant, mean[i] and
sigma should be logical, and all of the other nodes
should be stochastic. You change the node type by click on the
type field at the upper left part of the Doodle window.
When you change mean[i] to be logical, you should then enter its
formula into the field named value:.
Unlike in the model I presented in class, you should use an
exponential prior to ensure that alpha1 is positive and define
mean[i] using the formula mean[i] ~ alpha0 -
alpha1*sex[i]. As you create the model, you should ensure that
all of the parameters of each distribution are defined. The only
exception is that you should leave the lower bound and
upper bound parameters blank in all of the distributions.
When your model is finished, click on Doodle > Write Code. This will
open a window with the textual version of your model. Delete the
semicolon at the end of the first line in this file. I will call this
textual window the Text Model Window.
- Download the data file from here. Here is a brief
explanation of the notation.
- The notation
list(...) defines a list of variables
with values.
- The notation
c(a1,a2,a3...) defines a 1-dimensional
array whose contents are the values a1, a2,
and so on.
- Click on Model > Specification... to open the Specification
Tool.
- Double click on the word "model" at the start of the Text
Model Window. Then click on the "Check Model" button in the
Specification Tool. The words "model is syntactically correct" should
appear in the lower left edge of the main WinBUGS window.
- Now click on Open and open the data file. If WinBUGS asks
what kind of file it is, choose "text". Click on the word "list" at
the start of the data file window. Then click on the "Load Data"
button in the Specification Tool. The words "data loaded" should
appear in the lower left edge of the main WinBUGS window.
- Now edit the "num of chains" box in the Specification Tool to
have the value 2. Then click on the "Compile" button.
- Click on File > New to create a new empty window. In this
window, type a "list" command to define initial values for
alpha0, alpha1, and tau. Type
a second "list" command to define a second set of initial values for
these variables. Now double click on the first "list", move to the
Specification Tool and click on the "Load Inits" button. Notice that
the small box labeled "for chain" has the value 1, and that it
automatically increments to 2 when the initial values are loaded. The
words "chain initialized but other chain(s) contain uninitialized
values" should appear in the lower left edge of the main WinBUGS
window.
Now double click on the second "list" expression that you wrote, and
then click on the "Load Inits" button in the Specification Tool to
initialize the second chain. The words "model is initialized" should
appear in the lower left edge.
- Now we must tell WinBUGS what data to gather during the Markov
chain computation. Click on Inference > Samples... to open the Sample
Monitor Tool. Type the text "alpha0" into the "node" field and click
the "set" button. Repeat this for the "alpha1" and "tau" nodes. This
tells WinBUGS to remember each value of these variables that is
generated during the Gibbs sampling.
- We are now ready to run the chain. Click on Model >
Update... to open the Update Tool. The "updates" box specifies the
number of steps to run the Markov chain. The "refresh" box tells how
often to refresh any dynamic displays. Leave these unchanged and just
click on the "Update" button.
If all goes well, WinBUGS should rapidly perform the 1000 iterations.
However, if you get a run-time error, check to make sure that you have
the proper probability densities defined and that the initial values
make sense. I made the mistake of making tau a gaussian. This
allowed the variance of height[i] to become negative, and that caused
an error. Under Help > User Manual, there is a section on Tips and
Troubleshooting that provides some advice on how to fix problems. The
BUGS web page also has some pointers to other useful information.
- Now let's visualize what happened with the Markov chains. Go
to the Sample Monitor Tool. The box where you typed in the node names
has now become a drop-down menu. Select
alpha0 from this
menu and click on the "history" button. This will show the values of
alpha0 during the execution for both Markov chains. If
these values seem to be staying within the same range and that range
looks reasonable, this is comforting (but not particularly
illuminating). Click on the "quantiles" button, and you will see the
95% prediction interval as a function of time. This hopefully shows
the two Markov chains converging toward each other. You should
inspect this display for alpha1 and tau to
decide when the chains have converged. Suppose you decide that
they've converged after 400 iterations. Type 400 into the "beg" box
in the Sample Monitor Tool. Now any subsequent displays will ignore
the first 399 data points.
- The next step is to decide how frequently we can safely sample
from this markov chain to obtain approximately independent samples.
To do this, select a variable (e.g.,
alpha0) from the
node drop-down menu and click on the "auto cor" button. This displays
a bar graph showing the degree of autocorrelation between the sampled
value of alpha0 at time t and the sampled
values at times t - lag, for lag from 0 to 50. Suppose
you decide that there are clear autocorrelations up to lag 10. In
that case, type 10 into the "thin" box. This tells WinBUGS to compute
its statistics using 1 out of every 10 samples from the the Markov
chain. You can click "auto cor" again, and now you should see
significant autocorrelations only at lag 0.
With a convergence at iteration 400 and a thinning of 10, we will
have only 60 samples of each variable, so that may not been enough.
You can run the Markov chain for another 1000 steps by clicking on the
"Update" button in the Update tool. This will give us another 100
samples of each variable, which will give nicer estimates of the
posterior distributions.
- Finally, click on the "density" button to see the kernel
density estimate for each of the 3 nodes (
alpha0,
alpha1, and tau). Similarly, click on the
"stats" button to see the mean, median, and 95% prediction interval
for each parameter.
Running the Latent Variable Example
Now we will use WinBUGS to repat the latent variable example that I
did in class, where we only observe the height of 50 people but we
don't know their sexes.
- The only change you will need to make in the model is to make
sex[i] a stochastic node with the dbern
(Bernoulli) distribution, define a parent node theta to
specify the proportion parameter of the Bernoulli
distribution, and give theta a dbeta
distribution with parameters a=1 and b=1 (for a uniform prior).
- For the data file, you should remove the data for the
sex
variable from the "list".
- For the initial values, you will now need to provide initial
values for the
sex[i] variables and for
theta. For usex[i], I recommend tossing a
coin (or generating random numbers some way) and setting sex[i] to
either 0 or 1 (at random) separately for each i. You can
write your initial values as sex=c(0,1,1,0,1,...,).
- To monitor the values of the
sex variables, it is
impractical to use the Sample Monitor Tool. Instead, you should click
on Inference > Summary... to open the Summary Monitor Tool. Enter the
"sex" variable into the "node" field and click the "set" button. This
collects the statistics for sex[i] for each i, but it does not collect
the other more detailed information. After you have run the markov
chains, you can select "sex" from the "node" drop down menu in the
Summary Monitor Tool and then click on "Stats". One trick here is
that the data are collected from the start of the simulation, so you
can't tell WinBUGS when to start or what value of "thin" to use. To
accomplish this, you need to use the Update Tool. First, run the
Markov chains until they have converged. Then click "clear" on the
Summary Monitor Tool. Then go to the Update Tool and enter an
appropriate value for "thin". Now each update iteration will perform "thin"
number of actual updates and collect one sample. Hence, if you are
sampling every 10th value and you want 200 samples, you should set
"updates" to 200, and "thin" to 10. This will actually perform 2000
steps and draw 200 points. Now the Summary Monitor Tool will give you
good statistics. This is explained in the Tutorial section of Help >
User Manual.
Extending the Latent Variable Example (Optional)
If you have time and are curious, try to figure out how to extend
the latent variable model to have more than 2 possible sexes. You
should start by defining sex[i] to have a
dcat (categorical) distribution. A categorical
distribution is a multinomial distribution with K possible outcomes
(e.g., rolling a die with 6 possible outcomes). You can see an
example of this in the "Eyes" example from Help > Examples Volume II.
dcat requires a vector-valued parameter "proportions". This vector of
values is provided by a Dirichlet distribution ddirch.
The dirichlet distribution is the generalization of the beta
distribution to a vector of probabilities that must sum to 1. So
P[] ~ ddirch(alpha[]) where alpha =
c(1,1,1,1,1,1) would generate a vector of 6 probability values
that sum to 1.
The tricky part of having more than 2 sexes is that we can't just
use the model mean[i] = alpha0 - alpha1 * sex[i], because
we don't expect each sex k to be a constant amount taller than sex
k-1. One way to do this is to define a set of constant vectors
T1, T2, T3, T4,
T5 as follows:
list(
T1 = c(0, 1, 1, 1, 1, 1),
T2 = c(0, 0, 1, 1, 1, 1),
T3 = c(0, 0, 0, 1, 1, 1),
T4 = c(0, 0, 0, 0, 1, 1),
T5 = c(0, 0, 0, 0, 0, 1))
Now we can write the model as
mean[i] = alpha0 + alpha1*T1[sex[i]] + alpha2*T2[sex[i]] +
alpha3*T3[sex[i]] + alpha4*T4[sex[i]] + alpha5*T5[sex[i]]
In this case, alpha0 is the mean height of sex 1,
alpha0+alpha1 is the mean height of sex 2,
alpha0+alpha1+alpha2 is the mean height of sex 3, and so
on. I'm not convinced this is the best way to do this, because if the
Gibbs sampler increases alpha1, this causes all of the
remaining sexes to get taller so alpha2 then needs to be
decreased to compensate. But it does ensure that each sex is
well-defined (i.e., they are defined in increasing height order).
To fit more than 2 sexes, you will need more data. The following file
gives height data for all of the 21-25 year olds in the NIH study: height-21-25.txt. You will need to
reformat it for WinBUGS.
What to Turn In
For the two height models, you should be able to cut and paste the
Bayesian network and the relevant graphs into a Word file and then
print it out. I would like to see the following:
- The Doodle diagram
- The "history" plot for
alpha0 and your decision
about the number of iterations at which the chains have converged
- The "auto cor" plot for
alpha0 and your decision
about how often it is safe to sample from the chains
- The "density" plots for
alpha0, alpha1,
tau and theta (for the latent variable
model)
- The table of statistics for
sex[i] produced by the
Summary Monitor Tool (for the latent variable model). It would be
nice to convert this to a plot like the ones I showed in class.
Please give Bruce D'Ambrosio a hardcopy printout and email me the
Word file. I will be in Washington DC Tues-Wed-Thurs of next week, but
I'll look at them during my flight back across the country.
Tom Dietterich.